Trial Data Warehouse
Introduction
About
The protectr package serves as the R interface to the Protect Lab’s data warehouse. It allows you to access the extensive documentation of the warehouse, and to download and merge datasets locally via R.
Setup
To ensure data security, the warehouse follows a “two-factor” authentication procedure. First, a password is required to install the protectr package from a hidden GitHub repository. After the installation, you have full access to the trial documentation API (also accessible here). To download data from the warehouse directly into R, a further authentication step is required. All trial data is hosted on a secure PostgreSQL server at the Regional Computing Center Erlangen, which is part of the
FAU Erlangen-Nuremberg. To connect to the server, you must use the FAU VPN Client. FAU Staff can use their IDM credential to activate the VPN Client. External users should contact Mathias (mathias.harrer@fau.de) to request data exports or discuss VPN options.
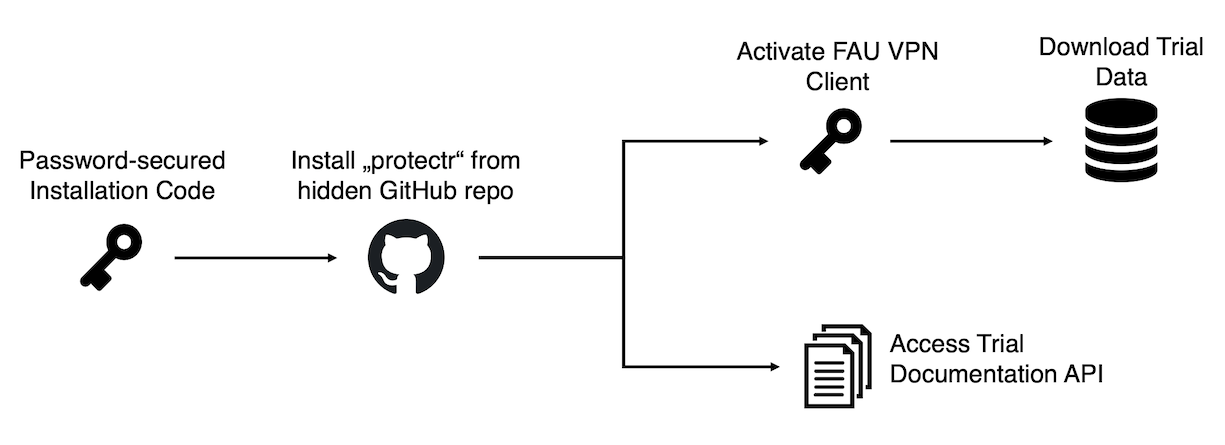
Once you were provided with the password, protectr can be downloaded from GitHub using this code:
if (!require("devtools")) {
install.packages("devtools")
}
devtools::install_github("MathiasHarrer/protectr",
build_vignettes = TRUE,
auth_token = "PASSWORD")If you are asked to update any existing packages on your computer during the installation, please choose “no”. If you run into trouble installing the package, please contact Mathias (mathias.harrer@fau.de).
After the installation, you can load protectr from your library.
library(protectr)The first step is to initiate the database. We do this using the get.db function, saving the output as db.
db <- get.db()Connected to warehouse succesfully.Documentation API
Examining the database object
The database object db now already contains the entire documentation. We can have a look at the upper-level objects using names.
names(db)[1] "Trials" "Outcomes" "Interventions" "Persons" "VariableDesc" We can also examine the object further, showing all trials included in the documentation by their name/shorthand.
db$Trials$Name stress_gui_264 stress_fod_263 diab_254 sleep_sjweh_128
"stress_gui_264" "stress_fod_263" "diab_254" "sleep_sjweh_128"
prevdep_406 prevdep_204 stress_studi_200 mdd_131
"prevdep_406" "prevdep_204" "stress_studi_200" "mdd_131"
stress_sh_264
"stress_sh_264" To get further information on a specific trial, we can use the get.trial.info function.
get.trial.info(db, trial = "stress_fod_263")Trial Info for: stress_fod_263
=======================
- Intervention: Get.On Fit im Stress
- Sample Size: 263
- Control Group Type: Waitlist
- Guidance Format: Feedback on Demand
- Primary Outcome: PSS-10
- Indication: Prevention - Selective
- No. of Trial Arms: 2
- Cutoff: PSS-10 >= 22
- Sample: General Adult
- Outcomes:
* sex
* age
* income
* ethnicity
* psychotherapy experience
* training experience
* relationship
* children
* employment
* education
* working years
* first-time helpseeker
* help-seeking attitudes
* BFI-10
* CES-D
* EQ5D
* ERI
* HADS-A
* ISI
* MBI-S Exhaustion
* PATHEV
* PSS-10
* PSWQ
* REQ
* SEK-27
* completed sessions
* SF-12
* UWES
* ZUF-8
- Timing Post: 7 weeks
- Timing FU1: 24 weeks
- Timing FU1: NA weeks
- Status: Completed
- Contact: Elena HeberTo only get the outcomes assessed in the trial (and included in the data warehouse), we can use the get.trial.outcomes function.
get.trial.outcomes(db, trial = "stress_fod_263")Baseline -----------------
$stress_fod_263
[1] "sex" "age"
[3] "income" "ethnicity"
[5] "psychotherapy experience" "training experience"
[7] "relationship" "children"
[9] "employment" "education"
[11] "working years" "first-time helpseeker"
[13] "help-seeking attitudes" "BFI-10"
[15] "CES-D" "EQ5D"
[17] "ERI" "HADS-A"
[19] "ISI" "MBI-S Exhaustion"
[21] "PATHEV" "PSS-10"
[23] "PSWQ" "REQ"
[25] "SEK-27" "completed sessions"
[27] "SF-12" "UWES"
[29] "ZUF-8"
Post ---------------------
$stress_fod_263
[1] "help-seeking attitudes" "CES-D" "ERI"
[4] "HADS-A" "ISI" "MBI-S Exhaustion"
[7] "PSS-10" "PSWQ" "REQ"
[10] "SEK-27" "completed sessions" "SF-12"
[13] "UWES" "ZUF-8"
FU1 ----------------------
$stress_fod_263
[1] "help-seeking attitudes" "CES-D" "ERI"
[4] "HADS-A" "ISI" "MBI-S Exhaustion"
[7] "PSS-10" "PSWQ" "REQ"
[10] "SEK-27" "completed sessions" "SF-12"
[13] "UWES" "ZUF-8" "EQ5D" We can also examine further what certain outcomes in a dataset mean, and for which characteristic certain factor levels may stand. We can use the get.var.description function to do this. We can access the documentation of either all outcomes in a trial, or only specific outcomes. Please note that outcomes which were assessed in several trials were always coded in the same way.
get.var.description(db, "prevdep_406", "inc") inc (factor)
income
1 = <10.000 EUR
2 = 10.000-30.000 EUR
3 = 30.000-40.000 EUR
4 = 40.000-50.000 EUR
5 = 50.000-60.000 EUR
6 = 60.000-100.000 EUR
7 = >100.000 EUR
8 = not statedIt is also possible to get a more general look on which outcomes where assessed in which trials. We can use the get.assessed.trials function to do this. We want to see in which trials PSS-10, CES-D, ERI and CSQ-8 (ZUF) data was assessed.
outcomes <- c("pss", "cesd", "eri", "zuf")
get.assessed.trials(db, outcomes) trial pss cesd eri zuf
1 stress_studi_200 1 1 0 0
2 stress_sh_264 1 1 1 1
3 stress_fod_263 1 1 1 1
4 stress_gui_264 1 1 1 1
5 prevdep_406 0 1 0 1
6 prevdep_204 0 1 0 0
7 diab_254 0 1 0 1
8 sleep_sjweh_128 0 1 1 1
9 sleep_hp_128 0 1 1 1
10 sleep_fod_128 0 1 1 1Access external trial documents
To attain more information, you can also use the get.registration function. It will open the online registration of the trial, if available, in your browser.
get.registration(db, trial = "prevdep_406")You can also access the published article as PDF online via get.trial.pdf.
get.trial.pdf(db, trial = "prevdep_406")Find optimal combination of studies to maximize outcome overlap
A common problem in analyzing cross-trial data is that outcomes are not consistently assessed. To facilitate finding clusters of studies with overlapping assessments, the get.maximum.coverage function. The function calls a biclustering algorithm (BiBit) internally and ranks the clusters it found. The algorithm can be run on the entire data warehouse, or a predefined selection of studies.
mc <- get.maximum.coverage(db, trials = c("stress_gui_264", "stress_fod_263",
"diab_254", "sleep_sjweh_128",
"prevdep_406", "prevdep_204",
"stress_studi_200", "mdd_131",
"stress_sh_264"))Transform matrix into arff format...DONE
BITPAT
============
ARFF file name: /var/folders/ny/__yp371n6bqdzh7y509mc5gc0000gp/T//RtmpRUP535/bibitdatadff8689c122.arff
Number of rows: 9
Number of columns: 83
Extra Columns Number: 6
The matrix is created
Number of biclusters at level 1: 21 in 240 milliseconds
Transforming into biclust output...DONE
=== RESULTS ==============================
Cluster 1 --------------------------------
- Studies included in cluster: 3
- Outcomes included in cluster: 29
- Total sample size: 791
Cluster 2 --------------------------------
- Studies included in cluster: 4
- Outcomes included in cluster: 18
- Total sample size: 1045
Cluster 3 --------------------------------
- Studies included in cluster: 4
- Outcomes included in cluster: 18
- Total sample size: 919
Cluster 4 --------------------------------
- Studies included in cluster: 4
- Outcomes included in cluster: 19
- Total sample size: 1197
Cluster 5 --------------------------------
- Studies included in cluster: 5
- Outcomes included in cluster: 16
- Total sample size: 1401
Cluster 6 --------------------------------
- Studies included in cluster: 4
- Outcomes included in cluster: 14
- Total sample size: 991
Cluster 7 --------------------------------
- Studies included in cluster: 4
- Outcomes included in cluster: 15
- Total sample size: 922
Cluster 8 --------------------------------
- Studies included in cluster: 2
- Outcomes included in cluster: 13
- Total sample size: 382
Cluster 9 --------------------------------
- Studies included in cluster: 2
- Outcomes included in cluster: 18
- Total sample size: 660
Cluster 10 --------------------------------
- Studies included in cluster: 3
- Outcomes included in cluster: 15
- Total sample size: 864
Cluster 11 --------------------------------
- Studies included in cluster: 2
- Outcomes included in cluster: 11
- Total sample size: 454
Cluster 12 --------------------------------
- Studies included in cluster: 2
- Outcomes included in cluster: 15
- Total sample size: 385
Cluster 13 --------------------------------
- Studies included in cluster: 2
- Outcomes included in cluster: 15
- Total sample size: 534
Cluster 14 --------------------------------
- Studies included in cluster: 6
- Outcomes included in cluster: 12
- Total sample size: 1529
Cluster 15 --------------------------------
- Studies included in cluster: 8
- Outcomes included in cluster: 8
- Total sample size: 1983
Cluster 16 --------------------------------
- Studies included in cluster: 6
- Outcomes included in cluster: 10
- Total sample size: 1456
Cluster 17 --------------------------------
- Studies included in cluster: 2
- Outcomes included in cluster: 21
- Total sample size: 610
Cluster 18 --------------------------------
- Studies included in cluster: 3
- Outcomes included in cluster: 11
- Total sample size: 810
Cluster 19 --------------------------------
- Studies included in cluster: 2
- Outcomes included in cluster: 18
- Total sample size: 537
Cluster 20 --------------------------------
- Studies included in cluster: 2
- Outcomes included in cluster: 20
- Total sample size: 335
Cluster 21 --------------------------------
- Studies included in cluster: 2
- Outcomes included in cluster: 11
- Total sample size: 331Let us explore Cluster 1, the optimal combination, a little further:
mc$ClusterInfo$Cluster1$studies[1] "stress_gui_264" "stress_fod_263" "stress_sh_264" mc$ClusterInfo$Cluster1$outcomes [1] "age" "BFI-10"
[3] "CES-D" "children"
[5] "completed sessions" "education"
[7] "employment" "EQ5D"
[9] "ERI" "ethnicity"
[11] "first-time helpseeker" "HADS-A"
[13] "help-seeking attitudes" "income"
[15] "ISI" "MBI-S Exhaustion"
[17] "PATHEV" "PSS-10"
[19] "PSWQ" "psychotherapy experience"
[21] "relationship" "REQ"
[23] "SEK-27" "sex"
[25] "SF-12" "training experience"
[27] "UWES" "working years"
[29] "ZUF-8" If you have questions concerning the protectr package, please contact Mathias at mathias.h.harrer@gmail.com.
Data Download
To download trial data from the data warehouse, you can use the get.data function. The functions allows to extract single trials, or retrieve a merged dataset containing several trials. When downloading more than one trial, you can decide if the resulting dataset should contain only the columns shared by all requested trials (merge = "intersection"), or all outcomes in all trials (merge = "union"). For the latter option, trials which did not assess a particular outcome have NA in respective column. Further options can be found by running ?get.data in your console.
# Select 3 trials by shorthand
trials <- c("prevdep_204", "prevdep_406", "mdd_131")
# Retrieve data
data <- get.data(trials, merge = "intersection", glimpse = T)Connecting to Protect RRZE DB...
SUCCESS
Observations: 741
Variables: 131
$ trial <chr> "prevdep_204", "prevdep_204", "prevdep_204", "prevdep_…
$ group <dbl> 1, 1, 1, 1, 0, 1, 1, 0, 1, 1, 0, 0, 0, 1, 0, 0, 1, 1, …
$ age <dbl> 45, 43, 30, 52, 26, 44, 36, 54, 37, 55, 34, 52, 56, 34…
$ sex <dbl> 0, 1, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, …
$ degree <dbl> 3, 3, 2, 2, 2, 2, 2, 3, 3, 3, 2, 2, 3, 3, 2, 2, 2, 3, …
$ rel <dbl> 1, 1, 0, 2, 0, 1, 1, 3, 0, 0, 1, 1, 2, 1, 1, 0, 1, 1, …
$ child <dbl> 1, 0, 1, 1, 0, 1, 1, 1, 0, 0, 1, 1, 1, 1, 0, 0, 1, 0, …
$ inc <dbl> 2, 2, 8, 3, 8, 2, 2, 2, 6, 1, 1, 4, 1, 8, 2, 3, 8, 2, …
$ employ <dbl> 1, 1, 1, 1, 0, 1, 1, 1, 1, 1, 1, 1, 1, 0, 1, 1, 1, 1, …
$ ethn <dbl> 0, 0, 5, 0, 0, 0, 0, 5, 0, 0, 5, 0, 5, 0, 0, 0, 5, 0, …
$ prevpsychoth <dbl> 1, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 1, 0, 0, 0, …
$ prevtraining <dbl> 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 1, 1, 1, 0, 0, 0, 0, …
$ depmed <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 1, …
$ atsphs.0.i1 <dbl> 2, 2, 0, 0, 3, 1, 3, 1, 0, 2, 1, 2, 2, 2, 3, 1, 0, 2, …
$ atsphs.0.i10 <dbl> 1, 1, 1, 1, 1, 1, 3, 0, 0, 1, 1, 0, 0, 1, 1, 2, 1, 1, …
$ atsphs.0.i2 <dbl> 1, 1, 2, 0, 0, 1, 0, 0, 1, 0, 1, 0, 0, 0, 0, 1, 1, 0, …
$ atsphs.0.i3 <dbl> 2, 1, 2, 3, 3, 2, 1, 2, 2, 2, 2, 0, 0, 2, 3, 2, 0, 2, …
$ atsphs.0.i4 <dbl> 2, 2, 1, 3, 1, 2, 1, 1, 1, 1, 2, 1, 0, 1, 1, 3, 3, 2, …
$ atsphs.0.i5 <dbl> 2, 2, 2, 1, 3, 1, 0, 2, 3, 3, 1, 3, 3, 2, 3, 1, 1, 3, …
$ atsphs.0.i6 <dbl> 1, 1, 2, 1, 3, 2, 1, 2, 3, 3, 1, 2, 2, 2, 2, 2, 1, 3, …
$ atsphs.0.i7 <dbl> 1, 1, 2, 2, 2, 2, 0, 1, 1, 2, 2, 2, 2, 2, 2, 2, 1, 1, …
$ atsphs.0.i8 <dbl> 2, 1, 1, 2, 0, 2, 2, 1, 1, 0, 2, 2, 1, 1, 0, 0, 2, 2, …
$ atsphs.0.i9 <dbl> 2, 0, 2, 2, 0, 1, 2, 1, 0, 1, 1, 1, 0, 0, 0, 2, 2, 2, …
$ atsphs.1.i1 <dbl> NA, 2, NA, NA, 3, 2, 3, 0, NA, 2, 2, 2, 3, 2, 3, 3, 0,…
$ atsphs.1.i10 <dbl> NA, 1, NA, NA, 0, 1, 2, 1, NA, 1, 2, 0, 0, 1, 1, 1, 3,…
$ atsphs.1.i2 <dbl> NA, 0, NA, NA, 0, 1, 0, 1, NA, 0, 0, 0, 0, 0, 0, 1, 1,…
$ atsphs.1.i3 <dbl> NA, 2, NA, NA, 3, 1, 3, 0, NA, 2, 2, 2, 3, 2, 3, 2, 2,…
$ atsphs.1.i4 <dbl> NA, 1, NA, NA, 1, 1, 0, 1, NA, 1, 1, 0, 3, 1, 0, 0, 3,…
$ atsphs.1.i5 <dbl> NA, 2, NA, NA, 2, 2, 2, 2, NA, 2, 2, 2, 3, 2, 3, 1, 2,…
$ atsphs.1.i6 <dbl> NA, 1, NA, NA, 2, 1, 1, 1, NA, 3, 2, 3, 3, 3, 2, 2, 0,…
$ atsphs.1.i7 <dbl> NA, 2, NA, NA, 2, 2, 2, 1, NA, 2, 2, 2, 3, 2, 2, 3, 1,…
$ atsphs.1.i8 <dbl> NA, 2, NA, NA, 0, 2, 2, 1, NA, 1, 1, 1, 0, 1, 0, 1, 2,…
$ atsphs.1.i9 <dbl> NA, 1, NA, NA, 0, 1, 1, 1, NA, 0, 2, 1, 0, 0, 0, 2, 3,…
$ badssf.0 <dbl> 21, 33, 23, 25, 37, 29, 30, 33, 8, 17, 26, 33, 35, 29,…
$ badssf.0.i1 <dbl> 1, 4, 2, 3, 2, 4, 2, 3, 0, 1, 2, 2, 2, 2, 2, 6, 4, 4, …
$ badssf.0.i2 <dbl> 6, 3, 1, 3, 3, 2, 6, 4, 0, 2, 3, 4, 6, 2, 2, 6, 0, 4, …
$ badssf.0.i3 <dbl> 3, 3, 3, 4, 4, 3, 3, 4, 0, 2, 2, 4, 3, 3, 3, 5, 2, 4, …
$ badssf.0.i4 <dbl> 1, 4, 3, 3, 4, 4, 0, 3, 0, 1, 5, 4, 2, 2, 1, 4, 2, 3, …
$ badssf.0.i5 <dbl> 2, 3, 2, 2, 5, 2, 6, 4, 2, 2, 3, 2, 3, 5, 4, 6, 4, 4, …
$ badssf.0.i6 <dbl> 1, 3, 3, 0, 3, 3, 0, 4, 0, 1, 4, 3, 4, 2, 0, 4, 0, 3, …
$ badssf.0.i7 <dbl> 2, 4, 4, 6, 6, 5, 6, 5, 2, 4, 2, 5, 6, 5, 1, 6, 0, 6, …
$ badssf.0.i8 <dbl> 3, 4, 2, 0, 4, 4, 1, 4, 0, 0, 1, 4, 5, 3, 2, 2, 4, 3, …
$ badssf.0.i9 <dbl> 2, 5, 3, 4, 6, 2, 6, 2, 4, 4, 4, 5, 4, 5, 2, 3, 3, 6, …
$ badssf.1 <dbl> NA, 34, NA, NA, 44, 37, 29, 31, NA, 19, 40, 36, 24, 20…
$ badssf.1.i1 <dbl> NA, 4, NA, NA, 4, 6, 1, 2, NA, 0, 5, 5, 1, 2, 3, 3, 5,…
$ badssf.1.i2 <dbl> NA, 4, NA, NA, 5, 4, 6, 4, NA, 2, 5, 4, 6, 2, 4, 6, 4,…
$ badssf.1.i3 <dbl> NA, 4, NA, NA, 5, 2, 1, 4, NA, 2, 5, 3, 2, 3, 4, 6, 4,…
$ badssf.1.i4 <dbl> NA, 4, NA, NA, 6, 5, 1, 3, NA, 2, 5, 3, 1, 1, 4, 5, 4,…
$ badssf.1.i5 <dbl> NA, 5, NA, NA, 5, 1, 5, 4, NA, 3, 5, 3, 1, 3, 4, 5, 5,…
$ badssf.1.i6 <dbl> NA, 2, NA, NA, 1, 6, 0, 4, NA, 1, 5, 5, 2, 0, 0, 1, 5,…
$ badssf.1.i7 <dbl> NA, 4, NA, NA, 6, 5, 6, 0, NA, 5, 5, 6, 4, 3, 4, 5, 6,…
$ badssf.1.i8 <dbl> NA, 4, NA, NA, 6, 5, 3, 6, NA, 1, 2, 3, 2, 3, 2, 6, 5,…
$ badssf.1.i9 <dbl> NA, 3, NA, NA, 6, 3, 6, 4, NA, 3, 3, 4, 5, 3, 3, 6, 2,…
$ badssf.2 <dbl> NA, 33, NA, NA, 41, 34, 30, 40, NA, 23, 48, 39, 30, 21…
$ badssf.2.i1 <dbl> NA, 3, NA, NA, 6, 4, 1, 3, NA, 1, 5, 5, 2, 2, 5, 6, 5,…
$ badssf.2.i2 <dbl> NA, 4, NA, NA, 6, 4, 6, 5, NA, 3, 6, 5, 5, 2, 5, 6, 3,…
$ badssf.2.i3 <dbl> NA, 4, NA, NA, 6, 4, 3, 5, NA, 3, 6, 4, 4, 3, 5, 6, 5,…
$ badssf.2.i4 <dbl> NA, 4, NA, NA, 4, 4, 1, 3, NA, 3, 5, 4, 4, 2, 5, 5, 4,…
$ badssf.2.i5 <dbl> NA, 4, NA, NA, 6, 3, 5, 5, NA, 3, 6, 3, 1, 2, 6, 5, 5,…
$ badssf.2.i6 <dbl> NA, 1, NA, NA, 2, 5, 1, 4, NA, 0, 5, 5, 2, 0, 4, 6, 5,…
$ badssf.2.i7 <dbl> NA, 5, NA, NA, 5, 5, 3, 6, NA, 5, 5, 5, 5, 4, 5, 6, 6,…
$ badssf.2.i8 <dbl> NA, 5, NA, NA, 2, 4, 5, 5, NA, 1, 5, 4, 4, 1, 6, 6, 5,…
$ badssf.2.i9 <dbl> NA, 3, NA, NA, 4, 1, 5, 4, NA, 4, 5, 4, 3, 5, 4, 2, 1,…
$ ceq.i1 <dbl> 6, 7, 7, 7, 6, 8, 2, 7, 8, 7, 5, 3, 5, 7, 7, 7, 5, 9, …
$ ceq.i2 <dbl> 4, 7, 5, 3, 6, 5, 2, 7, 7, 7, 5, 3, 5, 8, 7, 6, 5, 8, …
$ ceq.i3 <dbl> 4, 7, 2, 2, 9, 7, 2, 8, 6, 5, 5, 2, 5, 8, 9, 5, 1, 8, …
$ ceq.i4 <dbl> 34, 65, 34, 31, 70, 51, 0, 70, 69, 70, 20, 23, 65, 65,…
$ ceq.i5 <dbl> 4, 4, 4, 3, 5, 4, 2, 7, 6, 5, 5, 5, 5, 7, 5, 4, 1, 7, …
$ ceq.i6 <dbl> 33, 35, 25, 5, 40, 27, 8, 75, 55, 50, 20, 47, 63, 50, …
$ hadsa.0 <dbl> 9, 7, 11, 8, 6, 10, 6, 8, 16, 10, 15, 8, 11, 11, 13, 7…
$ hadsa.0.i1 <dbl> 1, 2, 2, 2, 1, 2, 2, 1, 3, 2, 3, 2, 3, 2, 3, 1, 2, 2, …
$ hadsa.0.i2 <dbl> 1, 1, 2, 1, 2, 2, 1, 2, 3, 0, 2, 1, 1, 2, 2, 0, 1, 2, …
$ hadsa.0.i3 <dbl> 2, 1, 2, 2, 0, 1, 1, 0, 3, 2, 3, 1, 1, 2, 2, 0, 1, 2, …
$ hadsa.0.i4 <dbl> 2, 2, 2, 1, 0, 1, 1, 2, 2, 2, 2, 1, 2, 2, 2, 2, 2, 1, …
$ hadsa.0.i5 <dbl> 1, 0, 1, 1, 1, 2, 1, 1, 2, 2, 1, 1, 1, 1, 2, 1, 1, 2, …
$ hadsa.0.i6 <dbl> 1, 1, 2, 0, 1, 1, 0, 2, 1, 0, 2, 2, 3, 1, 2, 2, 0, 0, …
$ hadsa.0.i7 <dbl> 1, 0, 0, 1, 1, 1, 0, 0, 2, 2, 2, 0, 0, 1, 0, 1, 1, 0, …
$ hadsa.1 <dbl> NA, 6, NA, NA, 5, 9, 5, 5, NA, 11, 10, 5, 12, 12, 11, …
$ hadsa.1.i1 <dbl> NA, 2, NA, NA, 1, 2, 1, 0, NA, 2, 1, 1, 3, 2, 3, 2, 1,…
$ hadsa.1.i2 <dbl> NA, 0, NA, NA, 2, 2, 1, 0, NA, 0, 2, 1, 2, 2, 2, 0, 1,…
$ hadsa.1.i3 <dbl> NA, 1, NA, NA, 0, 1, 2, 0, NA, 1, 1, 1, 1, 2, 2, 1, 1,…
$ hadsa.1.i4 <dbl> NA, 1, NA, NA, 0, 2, 1, 1, NA, 2, 1, 1, 3, 2, 2, 1, 1,…
$ hadsa.1.i5 <dbl> NA, 0, NA, NA, 1, 1, 0, 1, NA, 2, 1, 0, 1, 2, 1, 1, 1,…
$ hadsa.1.i6 <dbl> NA, 2, NA, NA, 1, 0, 0, 2, NA, 1, 2, 1, 2, 1, 1, 2, 0,…
$ hadsa.1.i7 <dbl> NA, 0, NA, NA, 0, 1, 0, 1, NA, 3, 2, 0, 0, 1, 0, 1, 0,…
$ hadsa.2 <dbl> NA, 5, NA, NA, 5, 9, 5, 8, NA, 11, 8, 7, 9, 14, 6, 10,…
$ hadsa.2.i1 <dbl> NA, 2, NA, NA, 1, 2, 1, 1, NA, 2, 1, 1, 2, 3, 1, 2, 1,…
$ hadsa.2.i2 <dbl> NA, 0, NA, NA, 0, 2, 2, 1, NA, 0, 1, 1, 2, 3, 1, 2, 1,…
$ hadsa.2.i3 <dbl> NA, 1, NA, NA, 0, 1, 1, 0, NA, 2, 2, 2, 1, 2, 1, 1, 1,…
$ hadsa.2.i4 <dbl> NA, 1, NA, NA, 1, 1, 0, 2, NA, 2, 1, 1, 2, 2, 1, 1, 1,…
$ hadsa.2.i5 <dbl> NA, 0, NA, NA, 1, 1, 1, 1, NA, 2, 1, 1, 0, 2, 1, 2, 1,…
$ hadsa.2.i6 <dbl> NA, 1, NA, NA, 1, 1, 0, 2, NA, 1, 1, 1, 2, 1, 1, 1, 0,…
$ hadsa.2.i7 <dbl> NA, 0, NA, NA, 1, 1, 0, 1, NA, 2, 1, 0, 0, 1, 0, 1, 0,…
$ spsi.npo.0 <dbl> 10, 6, 9, 3, 2, 10, 12, 1, 7, 6, 5, 3, 9, 19, 16, 1, 1…
$ spsi.npo.0.i10 <dbl> 2, 1, 2, 0, 0, 1, 0, 0, 3, 1, 2, 0, 2, 4, 4, 0, 4, 0, …
$ spsi.npo.0.i3 <dbl> 2, 3, 1, 3, 1, 3, 3, 1, 2, 1, 1, 1, 0, 3, 3, 1, 4, 2, …
$ spsi.npo.0.i5 <dbl> 2, 1, 3, 0, 1, 3, 4, 0, 0, 2, 1, 1, 3, 4, 3, 0, 2, 1, …
$ spsi.npo.0.i6 <dbl> 2, 0, 1, 0, 0, 2, 3, 0, 0, 1, 1, 1, 3, 4, 3, 0, 3, 0, …
$ spsi.npo.0.i8 <dbl> 2, 1, 2, 0, 0, 1, 2, 0, 2, 1, 0, 0, 1, 4, 3, 0, 1, 0, …
$ spsi.npo.1 <dbl> NA, 8, NA, NA, 2, 7, 18, 0, NA, 8, 7, 3, 8, 20, 13, 4,…
$ spsi.npo.1.i10 <dbl> NA, 2, NA, NA, 0, 1, 4, 0, NA, 2, 2, 1, 0, 4, 4, 0, 1,…
$ spsi.npo.1.i3 <dbl> NA, 2, NA, NA, 1, 2, 3, 0, NA, 1, 2, 1, 4, 4, 1, 4, 3,…
$ spsi.npo.1.i5 <dbl> NA, 1, NA, NA, 1, 1, 4, 0, NA, 2, 2, 1, 2, 4, 1, 0, 1,…
$ spsi.npo.1.i6 <dbl> NA, 2, NA, NA, 0, 2, 4, 0, NA, 1, 1, 0, 0, 4, 4, 0, 3,…
$ spsi.npo.1.i8 <dbl> NA, 1, NA, NA, 0, 1, 3, 0, NA, 2, 0, 0, 2, 4, 3, 0, 2,…
$ spsi.npo.2 <dbl> NA, 9, NA, NA, 3, 2, 11, 0, NA, 8, 10, 3, 5, 20, 13, 1…
$ spsi.npo.2.i10 <dbl> NA, 2, NA, NA, 0, 0, 2, 0, NA, 2, 2, 0, 1, 4, 3, 4, 2,…
$ spsi.npo.2.i3 <dbl> NA, 3, NA, NA, 2, 2, 3, 0, NA, 1, 2, 1, 2, 4, 4, 2, 3,…
$ spsi.npo.2.i5 <dbl> NA, 2, NA, NA, 1, 0, 3, 0, NA, 3, 2, 1, 0, 4, 3, 4, 1,…
$ spsi.npo.2.i6 <dbl> NA, 1, NA, NA, 0, 0, 1, 0, NA, 1, 2, 1, 1, 4, 1, 1, 4,…
$ spsi.npo.2.i8 <dbl> NA, 1, NA, NA, 0, 0, 2, 0, NA, 1, 2, 0, 1, 4, 2, 1, 1,…
$ spsi.ppo.0 <dbl> 9, 11, 5, 8, 15, 11, 6, 13, 9, 12, 14, 13, 15, 6, 8, 1…
$ spsi.ppo.0.i1 <dbl> 2, 2, 2, 1, 3, 3, 1, 3, 2, 3, 3, 3, 3, 2, 2, 3, 1, 2, …
$ spsi.ppo.0.i2 <dbl> 2, 3, 1, 1, 3, 3, 2, 3, 1, 2, 3, 2, 2, 1, 2, 2, 2, 2, …
$ spsi.ppo.0.i4 <dbl> 1, 2, 1, 3, 3, 2, 0, 3, 2, 1, 3, 3, 4, 1, 1, 1, 1, 2, …
$ spsi.ppo.0.i7 <dbl> 2, 3, 1, 3, 3, 2, 2, 2, 2, 3, 3, 3, 4, 1, 2, 2, 2, 3, …
$ spsi.ppo.0.i9 <dbl> 2, 1, 0, 0, 3, 1, 1, 2, 2, 3, 2, 2, 2, 1, 1, 2, 0, 2, …
$ spsi.ppo.1 <dbl> NA, 10, NA, NA, 12, 13, 8, 17, NA, 12, 14, 15, 12, 5, …
$ spsi.ppo.1.i1 <dbl> NA, 2, NA, NA, 3, 3, 2, 4, NA, 3, 3, 3, 3, 1, 2, 3, 2,…
$ spsi.ppo.1.i2 <dbl> NA, 2, NA, NA, 3, 3, 2, 3, NA, 2, 3, 3, 1, 2, 2, 2, 2,…
$ spsi.ppo.1.i4 <dbl> NA, 2, NA, NA, 2, 2, 0, 2, NA, 2, 2, 3, 2, 0, 1, 2, 2,…
$ spsi.ppo.1.i7 <dbl> NA, 3, NA, NA, 2, 3, 2, 4, NA, 2, 3, 3, 4, 1, 2, 1, 3,…
$ spsi.ppo.1.i9 <dbl> NA, 1, NA, NA, 2, 2, 2, 4, NA, 3, 3, 3, 2, 1, 1, 2, 2,…
$ spsi.ppo.2 <dbl> NA, 13, NA, NA, 17, 15, 8, 11, NA, 10, 13, 14, 16, 5, …
$ spsi.ppo.2.i1 <dbl> NA, 3, NA, NA, 4, 3, 2, 2, NA, 2, 3, 3, 3, 1, 3, 4, 2,…
$ spsi.ppo.2.i2 <dbl> NA, 3, NA, NA, 4, 3, 2, 2, NA, 2, 3, 3, 3, 1, 3, 4, 3,…
$ spsi.ppo.2.i4 <dbl> NA, 2, NA, NA, 3, 2, 1, 3, NA, 2, 2, 3, 4, 1, 0, 3, 2,…
$ spsi.ppo.2.i7 <dbl> NA, 3, NA, NA, 3, 4, 1, 2, NA, 2, 3, 3, 3, 1, 3, 4, 3,…
$ spsi.ppo.2.i9 <dbl> NA, 2, NA, NA, 3, 3, 2, 2, NA, 2, 2, 2, 3, 1, 1, 3, 0,…
$ sess <dbl> 0, 4, 2, 2, NA, 7, 7, NA, 0, 0, NA, NA, NA, 6, NA, NA,…